What is Proximity Ligation Assay?
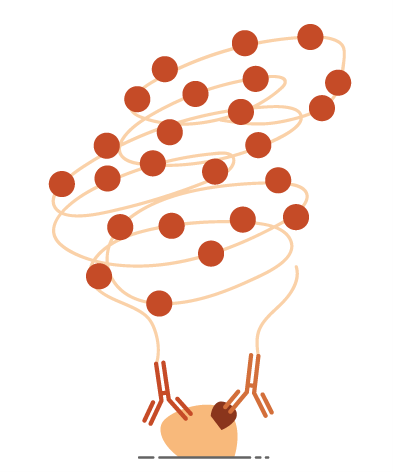
This method uses two oligo conjugated antibodies whose epitopes are located in close proximity to each other. To generate a signal, the oligos need to be able to form a DNA template that can be amplified in a rolling circle method to create a long string of single DNA that a complementary labelled oligo can bind to.

In this way the signal is ONLY generated where both antibodies have been bound closely enough for the oligos to bind. This spacing is around 40nm, which is below the resolution of even some of the best microscopes available today that are limited in their resolution due to the wavelength of light.
There are several ways that this technology can be used to advance spatial biology at present.
Where there is a need to demonstrate that proteins are binding to their ligands, such as with PD1 & PD/L1 immune checkpoint markers, antibodies can be chosen for each of the ligands and the signal is produced where the proteins are in conjunction with each other
To demonstrate where proteins are forming dimers, a monoclonal antibody can be separately labelled with the 2 oligo chains. As a monoclonal antibody should only recognise one epitope on the protein the binding of one part of the oligo should preclude the second binding and no signal is generated by a monomer – but it could bind to an adjacent protein where a dimer or multimer is being formed, thus enabling signal to be generated.
For post-translational modifications of proteins: As an example, phosphorylation of proteins can be hard to stain for easily. Although there are a few antibodies raised specifically against phosphorylated proteins, not all of them give satisfactory staining. Combining an antibody specific for the phosphorylation and another for the target of interest could offer a more flexible way to study these post translational modifications compared to a fridge full of phospho-target specific antibodies.
For particularly difficult antibodies for IHC, the use of PLA can offer a much cleaner signal as the density of off target binding needs to be higher to generate background signal – in a traditional ABC or polymer detection system, only 1 antibody aberrantly bound could contribute to background, whereas in a PLA method there would have to be 2 inappropriately bound antibodies very closely together to generate signal.
Developments
Although it’s great to be able to demonstrate where proteins are co-localizing, it can be useful to also see where each protein of interest is, as well as where they are interacting. There are new developments in this area with the Naveni® TriFlex Cell – which will yield one colour for protein ‘A’ another for protein ‘B’ and a third where there is ‘A+B’ together.
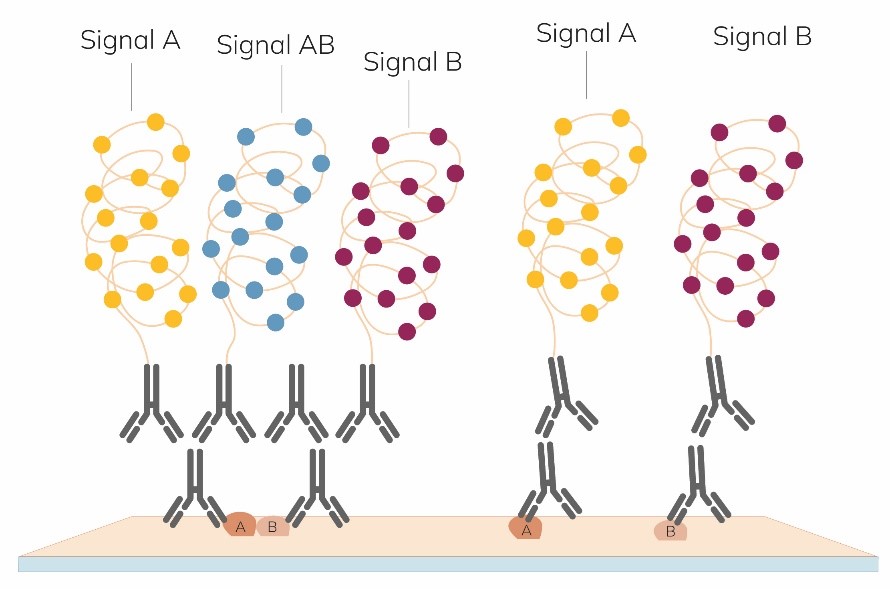
Once stained, sections or cell preparations can be interrogated for further epitopes, potentially with high plex-staining!
Previously, Proximity Ligation Assays have been done manually, but PLA kits for use on autostainers are now being developed.
The NaveniBright™ Bond RX HRP kits are the first approved kits for use on the LeicaBond RX/RXm autostainer that allow a fully automated in situ PLA assay to be run in under 10 hours.
There are currently 2 kits available, one specific for PD1/PD-L1 and a second flexible kit to run a PLA with a mouse and rabbit primary antibody pair.
Contact us to discuss your method and for assistance getting started!
| Selected kits from our preferred PLA kit manufacturer - Navinci Diagnostics |
|---|
| Naveni® Flex kits – for your own antibody pairs |
| Naveni® Phosphorylated Proteins |
| Naveni® Target specific kits |
| Naveni® Bond RX kits |
